Abstract
Soil pH conditions have important consequences for microbial community structure, their dynamics, ecosystem processes, and interactions with plants. Low soil pH affects the growth and functional activity of bacterial biocontrol agents which may experience a paradigm shift in their ability to act antagonistically against fungal phytopathogens. In this study, the antifungal activity of an acid-tolerant soil bacterium Bacillus amyloliquefaciens MBNC was evaluated under low pH and compared to its activity in neutral pH conditions. Bacterial supernatant from 3-day-old culture (approximately 11.2 × 108 cells/mL) grown in low pH conditions was found more effective against fungal pathogens. B. amyloliquefaciens MBNC harboured genes involved in the synthesis of secondary metabolites of which surfactin homologues, with varying chain length (C11–C15), were identified through High-Resolution Mass Spectroscopy. The pH of the medium influenced the production of these metabolites. Surfactin C15 was exclusive to the extract of pH 4.5; production of iturinA and surfactin C11 was detected only in pH 7.0, while surfactin C12, C13 and C14 were detected in extracts of both the pH conditions. The secretion of phytohormones viz. indole acetic acid and gibberellic acid by B. amyloliquefaciens MBNC was detected in higher amounts in neutral condition compared to acidic condition. Although, secretion of metabolites and phytohormones in B. amyloliquefaciens MBNC was influenced by the pH condition of the medium, the isolate retained its antagonistic efficiency against several fungal phyto-pathogens under acidic condition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant diseases caused by pathogens is an important factor limiting increase of crop yield. The intensive agricultural production system practiced globally relies on the use of agrochemicals for crop protection. Unfortunately, over-dependence and their indiscriminate use have led to the development of chemical resistant pathogens and impacted our environment by affecting non-target organisms and biodiversity. The pesticide residues left in the soil also make their way into the food chain system and affect human health (Zhang et al. 2011). The use of Microbial Biocontrol Agents (MBCAs) to control the infestation of crop pests and disease offers a sustainable alternative for crop production. The MBCAs exert their antagonistic effect by secreting inhibitory metabolites, competing with the pathogenic organism for nutrients or space, and induction of systemic resistance in plants (Saraf et al. 2014). However, the MBCAs released in the crop field encounter several abiotic stress conditions including low pH, salinity, temperature fluctuations, osmotic and oxidative stresses, availability of nutrients and water. These factors not only influence the growth and survivability of the applied MBCAs but also their antagonistic efficiency. The success of the MBCAs in suppressing the target pathogen is determined by its ability to tide over the stress condition while retaining its viability and efficacy. An understanding of the efficacy of the potential MBCAs in terms of its environmental fitness is thus important before they are released for commercial purpose. Very few studies have been directed towards understanding the effects of low pH condition on the effectiveness of biocontrol agents.
Soil acidity has its impact on several agricultural processes—including the influence of soil properties and processes that affect solubility and availability of nutrients for plant growth. Soil pH is regarded as the most important driver for soil microbial community structure and their function (Goswami et al. 2017). A shift from the neutral pH, either towards acidity or alkalinity, affects the growth and functionality of the soil microflora. Moreover, a neutral or near-alkaline soil pH promotes growth of bacteria while acidic pH favours fungal growth. The MBCAs inhabiting the soil are, therefore, likely to experience a paradigm shift in their ability to act antagonistically against certain phytopathogens depending on their ability to survive and supress the growth of the pathogen. Fungi adapt well in acidic condition and are reported to grow fivefold faster in acidic condition while the bacterial growth is inhibited under the same condition and decreases by a similar fold (Rousk et al. 2009). Under acidic conditions, the probability of the fungal pathogen becoming aggressive is high due to stifled activity of antagonistic bacteria. Low acidic soil condition (pH 4.5–5.5) is reported to suppress the growth and antagonistic activity of Bacillus cereus and Pseudomonas fluorescens while favouring the growth of the wilt causing pathogen, Ralstonia solanacearum (Li et al. 2017).
Previous studies have shown that the efficiency of plant growth promoting bacteria (PGPBs) decreases in acid stress conditions (Goswami et al. 2018). Therefore, an attempt was made to assess whether acidic conditions hinder the antagonistic efficiency of MBCAs. For carrying out this study, we took a previously isolated acid tolerant isolate of Bacillus amyloiquefaciens MBNC (Chowdhury et al. 2021) to evaluate its ability to act against six plant pathogenic fungi under acid stress condition. The spore-forming bacteria, B. amyloliquefaciens is widely distributed in nature and is well reported for its biocontrol activities against several pathogenic fungi viz., Alternaria panax, Botrytis cinerea, Colletotrichum acutatum, C. orbiculare, Corynespora cassicola, Fusarium oxysporum, Penicillium digitatum, Phytophthora capsici, Rhizoctonia solani, Stemphliyum lycopersici, Pyricularia grisea, Sclerotinia sclerotiorum (Ji et al., 2013). The bacteria exert its antagonistic effect mainly through the production of a broad range of antimicrobial compounds like lipopeptides, antibiotics, and siderophores (Zhang et al. 2016). The lipopeptides such as surfactin, iturin, and fengycin produced by the bacteria are reported to act antagonistically towards pathogens like Alternaria citri, Botryosphaeria sp., Colletotrichum gloeosporioides, Fusicoccum aromaticum, Lasiodiplodia theobromae, Penicillium crustosum, Phomopsis persea (Arrebola et al. 2010), Ralstonia solanacearum (Xiong et al. 2015), Rhizoctonia solani, Sclerotium rolfsii, Alternaria solani, Fusarium oxysporium (Jaivel et al. 2019). Additionally, B. amyloliquefaciens is also reported to promote plant growth through secretion of phytohormones, solubilising minerals and promoting soil aggregation (Deka et al. 2019).
Materials and methods
Fungal pathogens and their taxonomic characterization
Six phytopathogenic fungal isolates were collected from the Department of Plant Pathology, Assam Agricultural University, Assam which were identified as Pythium myriotylum 1NC, Fusarium verticilloides 2NC (synonym. Fusarium moniliforme), F. incarnatum 3NC, Athelia rolfsii 4NC (synonym. Sclerotium rolfsii), Fusarium oxysporum 5NC, and F. redolens 6NC. However, for further confirmation, the isolates were subjected to molecular characterization through sequencing of the ribosomal Internal Transcribed Spacer (ITS) region. The ITS region of the fungal genome was amplified with a pair of universal primer: ITS1 and ITS4 (Table 1). The PCR thermal profile was as follows: initial denaturation at 94 °C for 3 min; denaturation at 94 °C for 30 s (35 cycles), annealing at 50 °C for 30 s, extension at 72 °C for 45 s; and a final extension step at 72 °C for 7 min. The PCR amplicons were purified using GenElute™ PCR Clean-Up Kit (Sigma-Aldrich, USA) and the purified products were sequenced (Bio-Serve Biotechnologies, India). The sequence reads obtained post sequencing were assembled using CodonCode Aligner version 4.1 using default alignment parameters. The ends with low quality reads were trimmed and the generated contig for each sequence pair was saved as FASTA (.fasta) format for sequence analysis. A similarity search was employed using BLAST and compared with references at GenBank, NCBI to obtain their identity. Further, a phylogenetic tree was constructed from the ITS sequence data using MEGA 6.0. Sequences were aligned using clustalW and the Maximum Likelihood tree was prepared with Tamura-Nei model employing 1000 bootstrap replications.
Primary screening for antifungal activity of B. amyloliquefaciens MBNC
Dual culture disc diffusion method was used for confirming antifungal activity of the acid-tolerant isolate B. amyloliquefaciens MBNC. The culture plates were divided into two halves with a line through the diameter of each plate. Mycelial agar blocks (5-mm-diameter) of fungal phytopathogens were placed onto one half of the culture plate (15 mm away from the centre) and allowed to grow in individual plates for 2 days. The isolate B. amyloliquefaciens MBNC was inoculated in 50 ml Nutrient broth (NB) and incubated overnight at 37 °C, in shaking (180 rpm). This overnight grown culture was used for soaking sterile disc of filter paper (15 mm diameter), which were further placed at the opposite half of each culture plates containing the individual fungal pathogens. The plates were incubated at 28 °C until inhibition of fungal mycelia was observed.
Effect of pH on antagonistic activity of B. amyloliquefaciens MBNC
The effect of pH on the antifungal activity of B. amyloliquefaciens MBNC was assessed using dual culture method at pH 4.5 and pH 7.0. Spot inoculation of B. amyloliquefaciens MBNC was done onto one half of the culture plate, while the other half was inoculated with each of the fungal pathogens. For obtaining the desired neutral and acidic pH, the pH of the PDA medium was adjusted with sodium hydroxide and tartaric acid, respectively.
Preparation of bacterial supernatant
Bacillus amyloliquefaciens MBNC was inoculated in 50 ml Nutrient broth (NB) and incubated overnight at 37 °C, in shaking (180 rpm). One millilitre of the overnight culture was then used to inoculate fresh NB adjusted to pH 7.0 and pH 4.5 and incubated at 37 °C with shaking (180 rpm). For each pH, seven different NB was used as representative for 7 days (Day 1 to Day 7). Twenty millilitre culture of each pH was then transferred to a 50-ml tube, and the culture filtrate was obtained by centrifugation at 8000 rpm for 15 min at 4 °C. The supernatant was filtered through a 0.22-μm membrane filter into sterile tubes and kept at 4 °C until further use. This procedure was repeated consecutively for each day starting from Day 1 to Day 7.
Evaluation of antifungal activity
An aliquot of 100 μl sterile bacterial supernatant (previously stored in 4 °C), each of pH7.0 and pH 4.5 was embedded/spread on PDA plates and allowed to dry. This was followed by placing of a 5 mm-diameter mycelium agar block of each of the fungal isolates at the centre of individual plate and incubated in 28 °C incubator. The difference in fungal mycelia growth diameter at both pH condition was recorded consecutively for 7 days and calculated as inhibition percentage. The relative inhibition rate was calculated as follows:
Relative inhibition (%) = [(R1 − R2)/R1] × 100, where R1 is the colony diameter of control (fungal colony grown on PDA plates without the presence of any bacterial supernatant) and R2 is the diameter of fungal colony grown on PDA plate embedded with filter sterilized bacterial supernatant grown in either pH 7.0 or pH 4.5.
This procedure was repeated for the supernatant obtained from the cultures of Day 1 to Day 7. But since the bacterial supernatant obtained from the cultures grown for 3 days showed higher inhibition against all the fungal phytopathogen (data not shown), only the Day 3 supernatant has been taken into consideration for further studies.
Heat stability test
To evaluate the heat labile nature of the secondary metabolites, cell-free supernatant of B. amyloliquefaciens MBNC was exposed to 100 °C in a water bath for 1 h (Mariam et al. 2014). The cell-free supernatant was loaded into wells of PDA plates having 24-h-old culture of the fungal pathogens. Inhibition of mycelial growth near the punched well containing the heat-treated culture supernatant confirmed the heat stability of the antifungal compounds in the culture supernatant.
Detection of secondary metabolite biosynthetic genes
Eight genes viz. srfA (surfactin biosynthetic gene) dfnD (gene for difficidin biosynthesis), fenA (gene for fengycin biosynthesis), baeR (gene for bacillaene biosynthesis), ituA (gene for iturin A), bacD (gene for bacilysin biosynthesis), dhbE (gene for bacilibactin biosynthesis) and mlnA (gene involved in biosynthesis of macrolactins) belonging to the different secondary metabolite biosynthetic pathways were selected for screening through PCR based method. Genomic DNA from isolate B. amyloliquefaciens MBNC was used as template for PCR amplification of the secondary metabolite biosynthetic genes. The PCR reaction was performed using EmeraldAmp® GT PCR Master Mix (Takara, Japan) in 50 μl reaction volume containing 10 pmol of each primer (Table 1) and 50 ng of genomic DNA. The PCR condition was initial denaturation at 94 °C for 3 min; denaturation at 94 °C for 45 s (35 cycles), annealing temperature for 30 s, extension at 72 °C for 1.5 min; and a final extension step at 72 °C for 7 min. The amplified products were analysed on a 1.2% agarose gel.
Extraction of metabolites from culture supernatant of B. amyloliquefaciens MBNC
Preparation of bacterial supernatant was carried out using the procedure mentioned earlier (see “Preparation of bacterial supernatant”) followed by filtration through Whatman filter paper No. 2. Extraction of culture supernatant was carried out according to method described by Burianek and Yousef (2000) with slight modifications. The filtrate of the culture supernatant was acidified with 1 N HCl to pH 3.0. An equal volume of chloroform was mixed with the culture supernatant (1:1, v/v) followed by vigorous stirring for 20 min. The organic fraction that was soluble in chloroform was separated into a clean round bottom flask and concentrated under vacuum. This organic extract was used as crude extract and was dissolved in specific solvent according to requirements for further analysis. For confirming the antifungal activity of the extracted supernatant, the crude extract (extract of both pH 7.0 and pH 4.5) was dissolved in dimethyl sulphoxide (DMSO) and tested against the fungal pathogens by well diffusion method.
Identification and characterization of bioactive antifungal components produced under normal and acidic condition
To determine the extracellular active metabolites in the bacterial supernatant, the concentrated chloroform extract was re-dissolved in acetonitrile at a concentration of 100 μg/ml. The extract was then filtered through 0.22 μm PVDF membrane syringe filter (GE healthcare, USA) and 10 μl of filtrate was subjected to HRMS analysis in a Xevo G2-XS QT of HRMS system (Waters, USA). The positively ionized adduct ions of different compounds were calculated manually and compounds were identified by comparing them with the molecular information in PubChem database of NCBI (Maryland, USA).
Transcriptional regulation of srfA and ItuA under normal and acid stress conditions
Total RNA was extracted from B. amyloliquefaciens MBNC from cultures at pH 7.0 and pH 4.5, grown for 1 (Day 1) and 3 days (Day 3), using Purelink™ RNA mini kit (Ambion, Life Technologies, USA) following the manufacturer’s instructions. The first-strand cDNA synthesis was carried out using 2 μg total RNA in 50 μl reaction using Proto Script cDNA synthesis kit (Biolab, Life Technology) in a thermal cycler (ABI, USA). Based on the detection of bioactive metabolite in HR-MS analysis, expression profiles of the genes srfA and ituA were quantified by quantitative real-time PCR (qRT-PCR) in a QuantStudio 5 Real-Time PCR System (Applied Biosystems, USA) in a total reaction volume of 20 μL containing 10 nM of each primer and 50 ng cDNA templates. Primers used for real-time PCR study are listed in Table 1. The thermal program used was as follows: initial denaturation at 95 °C for 10 min followed by 35 cycles of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. The 16S rRNA gene was used as a reference gene (Goswami et al. 2018; Hazarika et al. 2019). The qRT-PCR runs were performed with three biological and technical replicates. The relative expression were evaluated using the 2−ΔΔCt method (Livak and Schmittgen 2001).
Evaluation of biocontrol potential of B. amyloliquefaciens MBNC
The effect of biocontrol potential of B. amyloliquefaciens MBNC was evaluated against six fungal phytopathogens during germination of bean (Phaseolus vulgaris) seeds. The bean seeds were surface sterilized with 1% sodium hypochlorite solution followed by three times washing with sterilized water. After that a suspension of bacterial cells was prepared in 0.85% saline solution and the cell numbers were adjusted to OD600 of 0.6 followed by soaking the bean seeds overnight in the bacterial suspension. For the control set, seeds were soaked overnight in 0.85% saline solution without any bacterial cells. The six fungal pathogens were cultured separately to prepare spore suspensions with approximately ~ 104 spores per ml as the initial concentration. The seeds (with and without bacterial treatment) were then exposed to each fungal spore suspensions separately for 30 min. A respective of eight seeds for each experimental condition were aseptically transferred to sterile petri plates lined with moistened filter paper. The experiment was carried out in triplicates. The germination percentage and root length were measured after 1 week of fungal treatment. Another plate containing surface sterilized seeds was kept as control to evaluate the normal germination efficiency of the seeds against the bacteria treated and untreated plates.
Screening plant growth promoting properties (PGP) of B. amyloliquefaciens MBNC
The bacterial isolates were screened for phytohormone production viz. Indole Acetic Acid (IAA) and Gibberellic Acid (GA) in-vitro. Quantitative estimation of IAA was performed by the method as described by Patten and Glick (2002). The isolate B. amyloliquefaciens MBNC was cultured overnight in DF salt minimal media (containing 4 g KH2PO4, 6 g Na2HPO4, 0.2 g MgSO4, 1 µg FeSO4, 10 µg H3BO3, 10 µg MnSO4, 70 µg ZnSO4, 50 µg CuSO4 and 10 µg MoO3 with 0.2% glucose, 0.2% gluconic acid and 0.2% citric acid/1 L) adjusted to pH 7.0 and pH 4.5. Twenty microlitres of the overnight-grown bacterial culture was transferred, respectively, into 5 ml of DF salt minimal media (adjusted to pH 7.0 and pH 4.5), amended with filter sterilized l-tryptophan (1000 μg/ml) and incubated at 30 °C for 72 h. This was followed by collecting the culture supernatant by centrifugation at 5000 rpm for 10 min. For measuring the amount of IAA produced, 1 ml of culture supernatant was mixed with 4 ml of Salkowski reagent (150 ml concentrated H2SO4, 250 ml of distilled H2O, 7.5 ml 0.5 M FeCl3∙6H2O solution). Development of pink colour indicates the production of IAA. After 20 min, optical density was taken at 535 nm by UV–VIS spectrophotometer (Spectroquant 300, Merck, Germany). Concentration of IAA produced by the bacterium was measured with the help of standard curve of IAA obtained in the range of 10–100 μ/ml. Quantitative estimation of GA was determined by the method described previously by Vikram et al. (2007). One ml of 0.6 OD fresh culture of B. amyloliquefaciens MBNC grown in pH 7.0 and pH 4.5 was inoculated in Nutrient broth amended with l-tryptophan (1 mg/ml) and incubated at 30 °C for 72 h. Five ml of the supernatant was taken in a test tube to which 0.4 ml of zinc acetate was added. After 2 min, 0.4 ml of potassium ferrocyanide was added and centrifuged at 1000 rpm for 15 min. To 3 ml of this supernatant, 3 ml of 30% HCl was added and incubated at 20 °C for 75 min. The blank sample was treated with 5% HCl and the absorbance of the sample as well as blank was measured at 254 nm in a spectrophotometer (Spectroquant 300, Merck, Germany). The procedure was carried out with B. amyloliquefaciens MBNC cultured in both pH 7.0 and pH 4.5. The amount of GA present in the extract was calculated from the standard curve of GA and expressed as μg/ml of the medium.
Statistical analysis
The graphs were prepared in Origin 8.5 and the data are represented as mean ± standard errors of three independent replications. Statistical comparisons between the two conditions (i.e., pH 7.0 and pH 4.5) were made using student’s t test in IBM SPSS version 25.0. The results were considered to be significant based on the corresponding p value (p < 0.05 or p < 0.01, whichever is applicable). Statistical significance between the data sets was designated as ‘*’ or ‘**’ based on p < 0.05 or p < 0.01, respectively.
Results
Identification of the fungal pathogens
Molecular characterization of the fungal isolates was carried out by sequencing the ITS regions followed by BLAST analysis to reveal all the six isolates as fungal phytopathogens. Based on the BLAST similarity scores, the identities of isolates were confirmed as Pythium myriotylum 1NC, Fusarium verticilloides 2NC (synonym. Fusarium moniliforme), Fusarium incarnatum 3NC, Athelia rolfsii 4NC (synonym. Sclerotium rolfsii), Fusarium oxysporum 5NC, and Fusarium redolens 6NC. The phylogenetic tree constructed using the ITS sequences of the fungal pathogens revealed their close evolutionary relationships with the type species (Supplementary Fig. S1). The ITS sequences were submitted to GenBank and the accession numbers obtained are given in Table 2.
Antagonistic activity of B. amyloliquefaciens MBNC against fungal phytopathogens
A primary screening by disc diffusion method was used to detect antifungal activity of the isolate B. amyloliquefaciens MBNC against the six different fungal plant pathogens. It was observed that isolate B. amyloliquefaciens MBNC could inhibit the mycelial growth of all the six fungal pathogens under the test condition (Supplementary Fig. S2). A comparative study was further carried out using dual culture technique at both pH 7.0 and pH 4.5 to evaluate the effect of low pH on the antagonistic activity of B. amyloliquefaciens MBNC. The bacterium was able to inhibit the fungal pathogens at both acidic and neutral pH conditions (Fig. 1). Highest inhibition of hyphal growth was recorded in case of Fusarium verticillioides, with approximately 94% and 82% inhibition at pH 7.0 and pH 4.5, respectively. The inhibitory activity was higher at pH 4.5 against F. oxysporum and A. rolfsii, as compared to that of pH 7.0 (Fig. 1). The effectiveness of bacterial culture supernatant collected from liquid culture with pH 4.5 and 7.0 in supressing fungal growth was also assessed. The bacterial supernatant of 3-day-old culture was found to be more effective in supressing the growth of the fungal pathogens in comparison to the 7-day culture supernatant as evident from the maximum inhibition percentage (Table 3). The 3-day-old bacterial supernatant from acidic pH was more effective against the fungal pathogens compared to that of neutral pH with an exception in F. verticillioides showing 25.13 ± 0.52% and 9.77 ± 0.38% inhibition in pH 7.0 and pH 4.5, respectively. In contrast, the 7-day old bacterial supernatant from acidic pH showed less efficiency against the fungal pathogens compared to that of neutral pH. Under the same condition, the inhibition was found to be negligible in case of A. rolfsii in both the pH (pH 7.0 and pH 4.5). The bacterial culture supernatant was ineffective against A. rolfsii due to the fast growth-rate of the fungus. In both the pH, the inhibition was found to be negligible in case of A. rolfsii. Initially, the inhibition zone was detected on the very next day of inoculation, which was subsequently replaced by the growing mycelia (Table 3).
Heat stability test of culture supernatant
The culture supernatant exposed to 100 °C did not lose its antagonistic activity as observed from the inhibition zone produced against each fungal phytopathogen (Supplementary table S1). Highest antagonistic activity was exhibited against P. myriotylum (with a zone diameter of 16 ± 0.67 mm and 14.33 ± 0.33 mm at days 3 and day 7, respectively) and F. redolens (zone diameter of 15.00 ± 0.00 mm and 13.67 ± 0.33 mm at days 3 and day 7, respectively) while lowest activity was observed against F. verticilloides (zone diameter of 3.00 ± 0.67 mm and 2.67 ± 0.33 mm at days 3 and day 7, respectively). This indicated that the bacterium produced certain antifungal compound(s) which remain active at high temperature.
Antifungal activity of metabolites produced by B. amyloliquefaciens MBNC
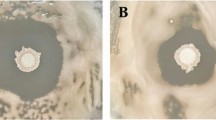
The DMSO solubilized chloroform extracts collected from both pH 7.0 and pH 4.5 showed significant antifungal activity as revealed by inhibition of fungal growth around the wells containing the extracts while, growth of the fungal cultures was not inhibited around the control (Fig. 2). The visual observation suggests that the chloroform extract of both pH 7.0 and pH 4.5 did not have any significant difference in terms of inhibition zone formation against the test fungal pathogens (Fig. 2a, b, d–f) except for the inhibition of Fusarium verticiloides by the extract obtained from the culture supernatant of pH 4.5 (Fig. 2c).
Well diffusion assay for detection of antifungal activity of chloroform extract of B. amyloliquefaciens MBNC at pH 4.5 and pH 7.0 against a Pythium myriotylum 1NC, b F. oxysporum 5NC, c F. verticilloides 2NC, d F. redolens 6NC, e Fusarium incarnatum 3NC and f Athelia rolfsii 4NC (7 indicates extract from pH 7.0, 4.5 indicates extract from pH 4.5; C indicates control wells with DMSO)
Detection of biosynthetic genes involved in secondary metabolite production
The genes involved in the synthesis of secondary metabolites in B. amyloliquefaciens MBNC was detected by PCR amplification with gene-specific primers. All the seven genes viz. srfA, dfnD, fenA, baeR, bacD, dhbE and mlnA showed amplification. The amplified products of size ~ 900 bp for dfnD, ~ 890 bp for srfA, ~ 960 bp for fenA, ~ 980 bp for ituA, ~ 670 bp for bacD, ~ 940 bp dhbE and ~ 920 bp for mlnA confirmed the presence of genes responsible for antifungal activity (Supplementary Fig. S3).
Determination of bioactive antifungal compounds produced by B. amyloliquefaciens MBNC
The chloroform extract of B. amyloliquefaciens MBNC supernatant was further subjected to the High-Resolution Mass Spectroscopy (HR-MS) analysis to detect and identify the metabolites aiding the biocontrol ability. The HR-MS analysis revealed the presence of surfactins with varying chain length including surfactin C12 (exact molecular mass 993.6 g/mol; retention time 3.97), surfactin C13 (exact molecular mass 1007.65 g/mol; retention time 4.123) and surfactin C14 (exact molecular mass 1021.66 g/mol; retention time 4.43) in the chloroform extract of both pH 7.0 and pH 4.5. Surfactin C15 (exact molecular mass 1035.68 g/mol; retention time 3.683) was exclusive to the extract of pH 4.5 with very low abundance; while production of surfactin C11 (exact molecular mass 979.60 g/mol; retention time 3.785) and iturin A (exact molecular mass 1042.54 g/mol; retention time 2.785) were only detected in pH 7.0 with low abundance (Fig. 3).
Detection of bioactive metabolites produced by B. amyloliquefaciens MBNC using HRMS. a Total Ion Chromatogram (TIC) of B. amyloliquefaciens MBNC extract grown at pH 7.0. b TIC of B. amyloliquefaciens MBNC extract grown at pH 4.5. c MS spectra of specific compounds detected in the TIC of extract collected from pH 7.0. d MS spectra of specific compounds detected in the TIC of extract collected from pH 4.5. The numbers mentioned in a, b denotes the compounds (i) iturin A (ii) surfactin C15 (iii) surfactin C11 (iv) surfactin C12 (v) surfactin C13 (vi) surfactin C14. The retention time for each compound is mentioned along with the spectra
Differential expression of srfA and ituA at pH 7.0 and pH 4.5 on Day 1 and Day 3
Quantitative real-time PCR analysis of srfA and ituA biosynthetic genes showed differential expression under acidic pH of 4.5. A significant difference in the expression of srfA at pH 4.5 was observed on day 1 but no significant difference was observed on day 3. However, ituA showed significant difference in its expression on both days at pH 4.5(Fig. 4). This indicated that pH of the medium had influence on the expression of the genes involved in surfactin and iturin biosynthesis. The similar expression pattern of srfA gene during day 3 in both acidic and neutral pH of bacterial culture confirmed the production of different surfactin derivatives in both the pH. However, the expression pattern of ituA gene did not correlate with the results of HR-MS analysis.
Effect of B. amyloliquefaciens MBNC on germination of seed of Phaseolus vulgaris infected with fungal phytopathogens
Bacillus amyloliquefaciens MBNC had a positive effect on the germination of bean (Phaseolus vulgaris) in terms of radicle formation, root elongation and suppression of fungal infection. The germination percentage and comparative root growth of bean under bacteria treated and untreated condition is shown in Table 4 and Fig. 5. The seeds that were pre-inoculated with bacterial suspension were able to counter the fungal phytopathogens and showed high germination efficiency. Although, there were no significant differences between bacteria treated and untreated seeds in terms of germination rate after 3 days of treatment (except for the seeds infected with F. oxysporum) (Fig. 5a), higher germination efficiency was observed after 7 days in bacteria-treated seeds due to notable reduction in the fungal infestation by most of the fungal pathogens (Fig. 5b). Control (uninfected) seeds exhibited ~ 100% germination efficiency regardless of bacterial treatment. Bacterial pre-treatment also generated pronounced effect on the root growth after seed germination. There was remarkable growth in the primary root length of bacteria-treated seeds compared to the bacteria-untreated seeds during fungal infestation (Fig. 5c–e). The differences were more pronounced after 7 days of treatment and even control (uninfected) seeds after bacterial treatment exhibited longer primary root length compared to the untreated seeds, indicating the bacterial influence over seed germination. An exception to this finding was found in case of A. rolfsii, where infection could not be controlled even with bacterial pre-treatment of the seeds which had low germination efficiency regardless of the treatment with bacteria.
Biocontrol efficiency of B. amyloliquefaciens MBNC against fungal infestation in germinating seeds of bean (Phaseolus vulgaris). a Germination percentage of bacteria treated and untreated seeds after 3 days of germination under fungal infestation; b germination percentage of bacteria treated and untreated seeds after 7 days of germination under fungal infestation; c average root length of germinated seeds after 3 days of germination; d average root length of germinated seeds after 7 days of germination; e photographic evidence showing the differences in the root length of bacteria treated and untreated seeds after 7 days of germination
Plant growth promoting properties (PGP) of B. amyloliquefaciens MBNC
The functional characteristics of B. amyloliquefaciens MBNC with regard to phytohormone (IAA and GA) production was evaluated both in neutral (pH 7.0) and acid-stress (pH 4.5) condition. The pH of the medium had an influence on the functionality of the isolate. Secretion of Indole-3-acetic acid (IAA) and gibberellic acid (GA) by the bacterium under neutral condition was higher (12.82 ± 0.12 µg/ml and 35.15 ± 0.005 µg/ml respectively) than in acidic condition (7.27 ± 0.06 µg/ml and 26.66 ± 0.003 µg/ml) (SupplementaryFig. S4). The secretion of phytohormone in acidic pH reduced significantly (p ≤ 0.05) compared to that of neutral pH.
Discussion
Several studies have reported the ability of Bacillus species to tolerate stress conditions such as high, acidity, alkalinity salinity or temperature (Niehaus et al. 1999; Horikoshi 2008). In this study, we took a previously isolated acid-tolerant bacterium B. amyloliquefaciens MBNC with plant growth-promoting ability to test for its antagonistic activity against plant fungal pathogens in acid-stress condition. B. amyloliquefaciens MBNC showed good antagonistic activity against P. myriotylum, F. incarnatum, F. redolens, F. verticilloides, F. oxysporum and S. rolfsii in acid-stress condition. The phytopathogenic fungi taken in this study are considered to be of significant threat to the agricultural crop production. The Genus Pythium comprises of several species that are mostly soilborne pathogens and cause serious economic loss on a wide variety by inhibiting seed germination, killing seedlings and reducing their vigour (Hendrix and Campbell 1973). Fusarium species belonging to a phytopathogenic group fungi cause wide range of plant diseases, such as crown rot, head blight, and scab disease on cereal grains, and vascular wilts on horticultural crops. Nearly all species of this genus produce mycotoxins that damage food and feed. The fungus Sclerotium rolfsii infects plant parts by causing damping-off of seedlings, stem canker, crown blight, root, crown, bulb, tuber and fruit rots. The mycelia of A. rolfsii spread rapidly and can remain active in soil for long period as sclerotia. The management of fungal diseases through microbial biological control agents is a safer alternative to the use of agrochemicals and more sustainable method of agricultural production. The antifungal activity of B. amyloliquefaciens MBNC ascertained by dual culture disc diffusion method revealed maximum inhibition by 3 days grown culture supernatant compared to 7 days grown culture. Incubation period of the bacterial culture has an effect on the production of metabolites. Bacteria produces secondary metabolites during its stationary phase when the growth gradually ceases but cells remain metabolically active. Therefore, bacterial culture with appropriate incubation time is crucial for proper detection of the metabolites. An early or a delayed sampling of the bacterial cultures might not be representative of the actual metabolite produced by the bacteria at a certain time point, as degradation, reutilization or conversion of the metabolite into another product may take place (Horak et al. 2019). In our study, B. amyloliquefaciens MBNC supernatant exhibited maximum inhibition on the 3rd day of incubation due to its maximum production of secondary metabolites which gradually decreased, with the minimum inhibition on day 7. B. subtilis produces active metabolites within the first 18 h which remain stable until the 96th hour (Marajan et al. 2018). B. amyloliquefaciensis well recognized as biocontrol agent and plant-growth promoting bacteria. The bacteria employ several strategies to act antagonistically against phytopathogens including secretion of lytic enzymes, production of antibiotics and volatile organic compounds and biosurfactants (Nunes 2012). Among the numerous antifungal compounds produced by Bacillus spp., lipopeptides belonging to the families of iturin, surfactin and fengycin are regarded as pivotal determinant of the biological control activity in B. amyloliquifaciens (Monteiro et al. 2016). These lipopeptides act on the fungal mycelial cell wall to inhibit conidial germination, causing morphological deformities and lysis of the mycelial cell wall leading to the death of the pathogen (Kumar et al. 2012). Several disease-causing plant pathogens including Pyricularia grisea, Sclerotinia sclerotiorum and Fusarium solani have been suppressed through application of B. amyloliquifaciens (Ji et al. 2013).
The organic fractions of the supernatant of B. amyloliquifaciens MBNC obtained from the chloroform extraction exhibited strong inhibition against the fungal isolates. An exception was observed in case of F. verticiloides where the chloroform extracts of the B. amyloliquifaciens MBNC grown in both pH conditions were less effective in inhibiting the fungal growth. The resistance of the pathogen may be attributed to the aggressive nature of the pathogen. Fusarium verticillioides (synonym, F. moniliforme) with variable aggressiveness has been reported from many plant species including maize (Leyva-Madrigal et al. 2017). The crude extract of the filtered supernatant of B. amyloliquefaciens obtained through chloroform extraction has been reported to act against several fungal pathogens (Kadaikunnan et al. 2015). Similarly, the culture supernatant of B. amyloliquefaciens DA12, extracted using butanol, was reported to completely suppress the germination of F. graminearum macroconidia when used at a concentration of 250 μg/ml (Lee et al. 2017). Previously, strong inhibition of fungal growth was also recorded in the chloroform extracted organic fractions of Bacillus subtilis SCB-1 (Hazarika et al., 2019).
The genes viz., srfA, dfnD, fenA, baeR, bacD, dhbE and mlnA, and ItuA involved in synthesis of secondary metabolite in the genome of B. amyloliquefaciens MBNC were detected by PCR. The presence of secondary metabolite gene clusters in Bacillus encoding for number of polyketides (PKs) viz. bacillaene, difficidin and macrolactins and lipopeptides (LPs) viz. iturins, fengycin, surfactin have been reviewed (Aleti et al. 2015). A study on whole genome mining estimated that 31% of the Firmicutes harbour non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKs) secondary metabolite gene clusters within their genomes. A majority (about 70%) of these bacteria encode NRPS while the rest encode PKs (Wang et al. 2014). The secondary metabolites. especially, PKs and LPs arm the bacteria with the ability to suppress disease in plants or activating plant immune response (Zhang et al. 2016). Antifungal potential of Bacillus subtilis SCB-1 against fungal phytopathogens showed presence of five genes involved in the synthesis of bioactive polyketides (bacillaene, difficidin and macrolactins) and lipopeptide (surfactin and fengycin) (Hazarika et al. 2019). Bacillaene has been reported to show antagonistic activity against bacteria and fungi (Patel et al. 1995; Chen et al. 2018). Difficidin and macrolactins comprises a group of broad-spectrum antibiotics produced by Bacillus sp. that functions by selective inhibition of protein synthesis (Schneider et al. 2007).
Further analysis to detect and determine the metabolites aiding in the biocontrol ability of B. amyloliquefaciens MBNC through HR-MS analysis revealed the presence of surfactins of different molecular weight and varying C-number (C11–C15). The surfactin C12, C13 and C14 homologues were abundantly detected in both pH 7.0 and pH 4.5, suggesting the direct role of these three molecules in controlling the growth of fungal pathogens. Earlier studies suggested that production of surfactin is favoured by pH nearing neutral or varying towards slightly acidic range (Monteiro et al. 2016). An increase in the production level of surfactin and fengycin by a factor of 3.7 and 2.13, respectively, on raising the pH level from 5.0 to 7.0 was reported earlier (Hmidet et al. 2017). Irrespective of the neutral or acidic pH, our findings suggested that surfactin variants were predominant in the culture supernatant of B. amyloliquefaciens MBNC. The antagonistic ability of our isolate to suppress fungal growth may be attributed to the production of surfactins in both pH conditions tested. The most commonly available surfactins in Bacillus species are surfactin C13, C14 and C15 (Bartal et al. 2018). Recent studies reported the biosynthesis of surfactin C11 and C12 (Sarwar et al. 2018; Bartal et al. 2018). The antifungal activity of surfactin is due to its interaction with the cell membrane and disruption of membrane stability (Carrillo et al. 2003). Iturins, fengycin, etc. are non-ribosomally synthesized lipopeptides that shows structural similarities to surfactin. Previous studies reported that some B. amyloliquefaciens and B. subtilis strains could co-produce fengycin, iturin and surfactin (Athukorala et al. 2009; Arrebola et al. 2010). In our study, HRMS analysis also detected iturinA in the supernatant of pH 7.0. The findings indicate that the production of the lipopeptides viz. surfactins of varying C-chain length and iturin A by B. amyloliquefaciens MBNC is effective in the suppression of the fungal pathogens. The decreased antifungal activity of the bacterial supernatant of pH 4.5 against F. verticilloides might be due to the absence of active iturin A in the chloroform extract of the bacterial culture grown in pH 4.5. Iturin A has been previously reported to govern the antifungal activity of Bacillus amyloliquefaciens DA12 against F. verticillioides and three other Fusarium species (Lee et al. 2017).
The results from HRMS analysis were further validated at transcript level through a qRT-PCR analysis to measure the expression level of srfA and ituA genes. The qRT-PCR analysis revealed increased expression of both the genes in Day 1 at pH 4.5. However, the transcript level of ituA on day 3 was higher at pH 4.5, while the expression level of srfA did not vary significantly on day 3 at pH 4.5 compared to the neutral pH. Biosynthesis of lipopeptides in Bacillus is regulated by complex metabolic network. Specific pleiotropic regulators including, the quorum sensing cluster comQXPA regulates biosynthesis of surfactins. Abiotic factors including temperature and pH of culture medium influence the expression of genes involved in the production of subtilosin A and iturin A in Bacillus strains (Mizumoto and Shoda 2007; Velho et al. 2011). Despite the abundance of ituA transcript level in both pH range, it was puzzling to note that HRMS analysis could not detect ituA at pH 4.5. This may be due to the regulatory steps leading to the final production of iturinA. Low pH of the medium affects the translation process of some genes (Goswami et al. 2018). It is also possible that techniques employed in the present study to detect the iturin may not have been sensitive enough for which more sensitive methods like Time of flight-secondary ion mass spectrometry (TOF–SIMS) may be required (Monteiro et al. 2016).
The efficiency of B. amyloliquefaciens MBNC in countering fungal infections during germination of bean seeds was established. Species of Pythium, Fusarium and Sclerotium are common seed-borne or soil-borne pathogens causing significant damages to common bean (Mahadevakumar et al. 2015; Binagwa et al. 2016; Marcenaro and Valkonen 2016). Three major pathogens from the genus Pythium viz. P. aphanidermatum, P. irregulare and P. myriotylum are reported to cause damping off disease of bean (Binagwa et al. 2016). Athelia rolfsii (synonym. Slerotium rolfsii) causes southern-blight with wilting and crown rot symptoms as well as leaf spot diseases of bean (Mahadevakumar et al. 2015). Likewise, several species of Fusarium cause wilt and root rot disease of bean, among which F. oxysporum, F. solani and F. incarnatum are most common (Marcenaro and Valkonen 2016). Experiment to evaluate the efficiency of B. amyloliquefaciens MBNC in supressing fungal pathogens during the germination of bean seeds showed considerable efficacy against the test pathogens (except A. rolfsii). B. amyloliquefaciens MBNC could not counter the A. rolfsi infection due to its rapid and aggressive growth over the seed. Researchers have previously reported the ability of B. amyloliquefaciens to inhibit pathogens in different vegetables including grey mould in tomato, scleotiorum rot in cucumber and powdery mildew in cucumber and pumpkin (Ji et al. 2013). The bacteria has also been used as biological control agents (BCA) to control green mould and blue mould rot on postharvest citrus (Yu 2009) and strawberry anthracnose (Essghaier et al. 2009). Iturin A produced by B. amyloliquefaciens effectively control Rhizoctonia solani have been reported earlier (Yu et al. 2002). In addition, soybean seeds coated with B. amyloliquifaciens were able to colonize the seedling rhizosphere and successfully control Rhizoctonia solani (Yu and Sinclair 1996).
The influence of pH on the plant growth promotion ability of different bacteria has been reported earlier (Goswami et al. 2018). The secretion of phytohormone in acidic pH reduced significantly (p ≤ 0.05) compared to that of neutral pH in this study. The adaptive mechanisms adopted by the bacteria under acid stress condition might have influenced the reduction in phytohormone secretion. Under acid-stress condition, diversion of more energy to survival mechanisms rather than metabolic processes like production of IAA or GA is imperative. Moreover, activity of enzymes involved in biosynthetic pathway involving phytohormone production may have been influenced by the pH of the medium, leading to the reduction in production of the same under acid stress.
Conclusion
This study established the potential of the isolate B. amyloliquefaciens MBNC as a promising candidate for biocontrol and plant growth promotion activities even under conditions of low pH. This isolate was found to be effective against several important fungal pathogens as it produces biometabolites such as surfactin and iturinA that aid in suppression of fungal phytopathogenic infections. Thus, this isolate is a promising biocontrol agent and can be effectively used against soil-borne fungal pathogen in acidic soil condition.
Availability of data and material
The ITS and 16S rRNA gene information is publicly available in the NCBI database.
References
Aleti G, Sessitsch A, Brader G (2015) Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J 13:192–203. https://doi.org/10.1016/j.csbj.2015.03.003
Arrebola E, Jacobs R, Korsten L (2010) Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J Appl Microbiol 108:386–395. https://doi.org/10.1111/j.1365-2672.2009.04438.x
Athukorala SNP, Fernando WGD, Rashid KY (2009) Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can J Microbiol 55:1021–1032. https://doi.org/10.1139/W09-067
Bartal A, Vigneshwari A, Bóka B et al (2018) Effects of different cultivation parameters on the production of surfactin variants by a Bacillus subtilis strain. Molecules 23:2675. https://doi.org/10.3390/molecules23102675
Binagwa PH, Bonsi CK, Msolla SN (2016) Evaluation of common bean (Phaseolus vulgaris) genotypes for resistance to root rot disease caused by Pythium aphanidermatum and Pythium splendens under screen house conditions. J Nat Sci Res 6:36–43
Burianek LL, Yousef AE (2000) Solvent extraction of bacteriocins from liquid cultures. Lett Appl Microbiol 31:193–197. https://doi.org/10.1046/j.1365-2672.2000.00802.x
Carrillo C, Teruel JA, Aranda FJ, Ortiz A (2003) Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochem Biophys Acta 1611(1–2):91–97. https://doi.org/10.1016/s0005-2736(03)00029-4
Chen K, Tian Z, Luo Y et al (2018) Antagonistic activity and the mechanism of Bacillus amyloliquefaciens DH-4 against citrus green mold. Phytopathology 108:1253–1262. https://doi.org/10.1094/PHYTO-01-17-0032-R
Chowdhury N, Goswami G, Boro RC, Barooah M (2021) A pH-dependent gene expression enables Bacillus amyloliquefaciens MBNC to adapt to acid stress. Curr Microbiol 78:3104–3114. https://doi.org/10.1007/s00284-021-02573-y
Deka P, Goswami G, Das P et al (2019) Bacterial exopolysaccharide promotes acid tolerance in Bacillus amyloliquefaciens and improves soil aggregation. Mol Biol Rep 46:1079–1091. https://doi.org/10.1007/s11033-018-4566-0
Essghaier B, Fardeau ML, Cayol JL et al (2009) Biological control of grey mould in strawberry fruits by halophilic bacteria. J Appl Microbiol 106:833–846. https://doi.org/10.1111/j.1365-2672.2008.04053.x
Goswami G, Deka P, Das P et al (2017) Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech. https://doi.org/10.1007/s13205-017-0864-9
Goswami G, Panda D, Samanta R et al (2018) Bacillus megaterium adapts to acid stress condition through a network of genes: insight from a genome-wide transcriptome analysis. Sci Rep 8:16105. https://doi.org/10.1038/s41598-018-34221-0
Hazarika DJ, Goswami G, Gautom T et al (2019) Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol 19:71. https://doi.org/10.1186/s12866-019-1440-8
Hendrix FF, Campbell WA (1973) Pythiums as plant pathogens. Annu Rev Phytopathol 11:77–98. https://doi.org/10.1146/annurev.py.11.090173.000453
Hmidet N, Ben Ayed H, Jacques P, Nasri M (2017) Enhancement of surfactin and fengycin production by Bacillus mojavensis A21: application for diesel biodegradation. Biomed Res Int 2017:1–8. https://doi.org/10.1155/2017/5893123
Horak I, Engelbrecht G, van Rensburg PJ, Claassens S (2019) Microbial metabolomics: essential definitions and the importance of cultivation conditions for utilizing Bacillus species as bionematicides. J Appl Microbiol 127(2):326–343. https://doi.org/10.1111/jam.14218
Horikoshi K (2008) Past, present and future of extremophiles. Extremophiles 12:1–2. https://doi.org/10.1007/s00792-007-0127-5
Jaivel N, Ramasamy R, Sivakumar U, Marimuthu P (2019) Antimicrobial activity and spectroscopic characterization of surfactin class of lipopeptides from Bacillus amyloliquefaciens SR1. Microb Pathog. https://doi.org/10.1016/j.micpath.2019.01.037
Ji SH, Paul NC, Deng JX et al (2013) Biocontrol activity of Bacillus amyloliquefaciens CNU114001 against fungal plant diseases. Mycobiology 41:234–242. https://doi.org/10.5941/MYCO.2013.41.4.234
Kadaikunnan S, Rejiniemon T, Khaled JM et al (2015) In-vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann Clin Microbiol Antimicrob 14:9. https://doi.org/10.1186/s12941-015-0069-1
Kumar P, Dubey RC, Maheshwari DK (2012) Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res 167:493–499. https://doi.org/10.1016/J.MICRES.2012.05.002
Lee T, Park D, Kim K et al (2017) Characterization of Bacillus amyloliquefaciens DA12 showing potent antifungal activity against mycotoxigenic Fusarium species. Plant Pathol J 33:499–507. https://doi.org/10.5423/PPJ.FT.06.2017.0126
Leyva-Madrigal KY, Sandoval-Castro E, Calderón-Vázquez CL et al (2017) Pathogenic and genetic variability of Fusarium verticillioides from maize in northern Mexico. Can J Plant Pathol 39:486–496. https://doi.org/10.1080/07060661.2017.1378726
Li S, Liu Y, Wang J et al (2017) Soil Acidification aggravates the occurrence of bacterial wilt in South China. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00703
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mahadevakumar S, Tejaswini GS, Janardhana GR, Yadav V (2015) First report of Sclerotium rolfsii causing southern blight and leaf spot on common bean (Phaseolus vulgaris) in India. Plant Dis 99:1280–1280. https://doi.org/10.1094/PDIS-01-15-0125-PDN
Marajan C, Alias S, Ramasamy K, Abdul-Talib S (2018) The effect of incubation time, temperature and pH variations on the surface tension of biosurfactant produced by Bacillus spp. AIP conference. AIP, p 020047
Marcenaro D, Valkonen JPT (2016) Seedborne pathogenic fungi in common bean (Phaseolus vulgaris cv. INTA Rojo) in Nicaragua. PLoS ONE 11:e0168662. https://doi.org/10.1371/journal.pone.0168662
Mariam SH, Zegeye N, Tariku T et al (2014) Potential of cell-free supernatants from cultures of selected lactic acid bacteria and yeast obtained from local fermented foods as inhibitors of Listeria monocytogenes, Salmonella spp. and Staphylococcus aureus. BMC Res Notes 7:606. https://doi.org/10.1186/1756-0500-7-606
Mizumoto S, Shoda M (2007) Medium optimization of antifungal lipopeptide, iturin A, production by Bacillus subtilis in solid-state fermentation by response surface methodology. Appl Microbiol Biotechnol 76:101–108. https://doi.org/10.1007/s00253-007-0994-9
Monteiro FP, de Medeiros F, Henrique Vasconcelos Ongena M, Franzil L, de Souza P, Estevão de Souza JT (2016) Effect of temperature, pH and substrate composition on production of lipopeptides by Bacillus amyloliquefaciens 629. African J Microbiol Res 10:506–1512
Niehaus F, Bertoldo C, Kähler M, Antranikian G (1999) Extremophiles as a source of novel enzymes for industrial application. Appl Microbiol Biotechnol 51:711–729. https://doi.org/10.1007/s002530051456
Nunes CA (2012) Biological control of postharvest diseases of fruit. Eur J Plant Pathol 133:181–196. https://doi.org/10.1007/s10658-011-9919-7
Patel PS, Huang S, Fisher S et al (1995) Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: production, taxonomy, isolation, physico-chemical characterization and biological activity. J Antibiot (tokyo) 48:997–1003
Patten CL, Glick BR (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68:3795–3801. https://doi.org/10.1128/AEM.68.8.3795-3801.2002
Rousk J, Brookes PC, Bååth E (2009) Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol 75:1589–1596. https://doi.org/10.1128/AEM.02775-08
Saraf M, Pandya U, Thakkar A (2014) Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol Res 169:18–29. https://doi.org/10.1016/j.micres.2013.08.009
Sarwar A, Nadeem M, Imran M et al (2018) Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol Res 209:1–13. https://doi.org/10.1016/j.micres.2018.01.006
Schneider K, Chen X-H, Vater J et al (2007) Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod 70:1417–1423. https://doi.org/10.1021/np070070k
Velho RV, Caldas DGG, Medina LFC et al (2011) Real-time PCR investigation on the expression of sboA and ituD genes in Bacillus spp. Lett Appl Microbiol 52:660–666. https://doi.org/10.1111/j.1472-765X.2011.03060.x
Vikram A, Hamzehzarghani H, Alagawadi AR et al (2007) Production of plant growth promoting substances by phosphate solubilizing bacteria isolated from vertisols. J Plant Sci 2:326–333. https://doi.org/10.3923/jps.2007.326.333
Wang H, Fewer DP, Holm L et al (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci 111:9259–9264. https://doi.org/10.1073/pnas.1401734111
Xiong H, Li Y, Cai Y, Cao Yu, Wang Y (2015) Isolation of Bacillus amyloliquefaciens JK6 and identification of its lipopeptides surfactin for suppressing tomato bacterial wilt. RSC Adv 5(100):82042–82049. https://doi.org/10.1039/C5RA13142A
Yu S (2009) Biological control of postharvest green and blue mold rots of citrus fruits by Bacillus amyloliquefaciens BCL251. Chungnam National University, Daejeon
Yu GY, Sinclair JB (1996) Evaluation of Bacillus amyloliquefaciens B94 for control of Rhizoctonia seedling disease on soybeans. Phytopathology 86:S54
Yu G, Sinclair J, Hartman G, Bertagnolli B (2002) Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem 34:955–963. https://doi.org/10.1016/S0038-0717(02)00027-5
Zhang W, Jiang F, Ou J (2011) Global pesticide consumption and pollution: with China as a focus. Proc Int Acad Ecol Environ Sci 1:125–144
Zhang S, Jiang W, Li J et al (2016) Whole genome shotgun sequence of Bacillus amyloliquefaciens TF28, a biocontrol entophytic bacterium. Stand Genom Sci. https://doi.org/10.1186/s40793-016-0182-6
Acknowledgements
The authors wish to acknowledge the Department of Biotechnology (DBT), Government of India, for financial support for the Project, “Screening of soil microbes for acid tolerance gene” under DBT-North East Centre for Agricultural Biotechnology (DBT-NECAB), Assam Agricultural University, Jorhat, India. The authors are thankful to Dr. M. K. Modi, Head, Department of Agricultural Biotechnology and Dr. B. K. Sarmah, Director DBT-AAU Centre, AAU, Jorhat for providing the requisite facilities and recommendations. The authors gratefully acknowledge the Department of Plant Pathology, Assam Agricultural University, Jorhat, India for providing the fungal isolates.
Funding
This study was supported by funds received from the Department of Biotechnology (DBT), Government of India for the project “Screening of soil microbes for acid tolerance gene” under DBT-NECAB, Assam Agricultural University (AAU), Jorhat, India.
Author information
Authors and Affiliations
Contributions
Conceptualization and Methodology: MB, GG, NC; Formal analysis and investigation: NC, GG, DJH, US, SB; Data Curation: NC, GG, DJH; Writing—original draft preparation: NC; Writing—review and editing: NC, GG, DJH, RCB, MB; Resources: MB, RCB; Supervision: MB.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the authors have given consent for publication.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chowdhury, N., Hazarika, D.J., Goswami, G. et al. Acid tolerant bacterium Bacillus amyloliquefaciens MBNC retains biocontrol efficiency against fungal phytopathogens in low pH. Arch Microbiol 204, 124 (2022). https://doi.org/10.1007/s00203-021-02741-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02741-5